Diagnosis of African Swine Fever
Discover our magnetic virus capture & DNA extraction solutions

Disease profile
African Swine Fever (ASF) is a hemorrhagic fever in pigs. It is caused by the ASF Virus, an enveloped virus with an almost 200kb long double-strand DNA genome. It infects wild pigs as well as domestic pigs. Natural reservoirs are boars and warthogs but also soft ticks of the genus Ornithodoros. ASF is neither infectious nor dangerous to humans.
African Swine Fever is highly contagious and can be transmitted through direct contact with infected animals and their body fluids & feces, but also through contaminated feed. In nature, blood meals of ticks do also play a role in the transmission. Infected animals do not show severe symptoms in the first days after infection. Yet, the symptoms are generally similar to those of "classical" Swine Fever. However, more virulent strains cause a striking change in behavior and the pigs show blue-purple coloration on their skin caused by the hemorrhagic activitiy of ASFV. Animals infected with rather mild strains of ASF virus may lose weight and show other unspecific signs.
African Swine Fever Virus replicates in macrophages, whose cellular processes are severely modulated by the infection. The virus also interfers with important signalling pathways and the immune response by macrophages. The first outbreak of African Swine Fever was reported in Kenya in the early 20th century after settlers started mass pig farming in Africa. The disease remained a local problem for about 50 years, before it started to spread into other regions. In recent years, ASF spreaded remarkably into new countries around the world. Meanwhile, it has been reported in over 70 countries, mainly in Africa, East Asia and Europe.
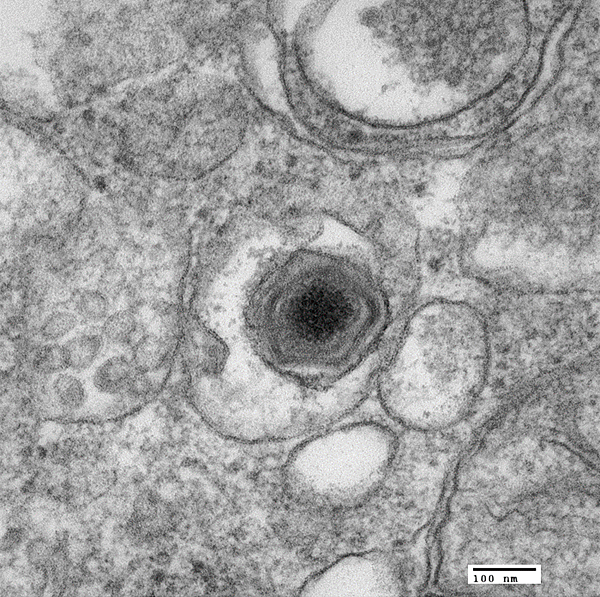
Molecular diagnosis
African Swine Fever is usually diagnosed and distinguished from classical swine fever by using either methods such as ELISA or through molecular detection. Typically, molecular diagnosis is performed using blood and organ tissue samples as specimens. In countries with a more preventive and surveilling ASF strategy like China, further non-invasive sample types are used, e.g. swine oral fluid, nasal swabs and even environmental swabs from surfaces.
Molecular diagnosis of African Swine Fever requires a sensitive and cost-efficient extraction of viral DNA. Ideally, the DNA extraction protocol can be automated, because at outbreaks as well as in surveillance programs the number of samples is rather high. Amplification and detection of ASFV DNA itself is usually performed using real-time PCR, but also through isothermal amplification methods like RPA, RAA or LAMP.
Applicable Xpedite Diagnostics products
SwiftX™ Hi-Sense (25 extractions)
For magnetic bead-based capture of ASF viral particles and rapid extraction of ASFV DNA.
Validated for oral & nasal swabs, swine oral fluid, and environmental samples.
SwiftX™ DNA (25 extractions)
For magnetic bead-based capture of ASF viral particles and rapid extraction of ASFV DNA.
Validated for whole blood, tissue samples, oral & nasal swabs, swine oral fluid, and environmental samples.
Go to productUser protocols
Magnetic capturing of ASF virus particles from swine oral fluid (SOF) and nasal/oral swab eluates followed by DNA extraction (SwiftX™ Hi-Sense)
Beijing UniScience, laboratory validation
- 1000µL SOF or swab eluate (in PBS or water) were taken for DNA extraction
- 700µL Buffer EN and 30µL Beads B were added and mixed
- mixture was incubated for 3 minutes for binding of ASFV particels and macrophages to Beads B
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads B with ASFV particles were resuspended in 75µL Buffer DL
- mixture was heated for 15 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time PCR detection
Magnetic capturing of ASF virus particles from tail root blood swabs and environmental swab eluates followed by DNA extraction (SwiftX™ Hi-Sense)
Beijing UniScience, laboratory validation
- 1000µL swab eluate (in PBS or water) were taken for DNA extraction
- 1000µL Buffer EN and 30µL Beads B were added and mixed
- mixture was incubated for 3 minutes for binding of ASFV particels and macrophages to Beads B
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads B were resuspended in 500µL Buffer EN
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads B with ASFV particles were resuspended in 75µL Buffer DL
- mixture was heated for 15 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time PCR detection
Magnetic capturing of ASF virus particles from whole blood followed by DNA extraction (SwiftX™ DNA)
Xpedite Diagnostics, laboratory validation
- 200µL EDTA whole blood were taken for DNA extraction
- 400µL Buffer EN and 30µL Beads A were added and mixed
- mixture was incubated for 3 minutes for binding of ASFV particels and macrophages to Beads A
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A were resuspended in 500µL Buffer EN
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A with ASFV particles were resuspended in 100µL Buffer DL
- mixture was heated for 15 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time PCR detection
Magnetic capturing of ASF virus particles from homogenized spleen followed by DNA extraction (SwiftX™ DNA)
Xpedite Diagnostics, laboratory validation
- 200µL spleen homogenate were taken for DNA extraction
- 400µL Buffer EN and 30µL Beads A were added and mixed
- mixture was incubated for 3 minutes for binding of ASFV particels and macrophages to Beads A
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A were resuspended in 500µL Buffer EN
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A with ASFV particles were resuspended in 100µL Buffer DL
- 5µL Proteinase K was added and mixed
- mixture was heated for 5 minutes at 60°C
- mixture was heated for 15 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time PCR detection
Technical information
Documents and Videos
Scientific literature
References
There are currently no scientific publications related to the use of our extraction kits for ASFV detection available.