Diagnosis of intestinal and urinary Schistosomiasis
Discover our magnetic cell capture & DNA extraction solution for urogenital and intestinal schistosomiasis

Disease profile
Schistosomiasis belongs to the group of Neglected Tropical Diseases (NTDs). It is also called Bilharzia or Snail fever. The disease is caused by infection with Schistosoma trematodes (also called blood flukes or flat worms). The infection happens during contact of people with infested fresh water, such as children playing in water ponds and farmers or fishermen working on fields and in lakes, respectively. The infestation of these water reservoirs occurs through release of worm eggs by fresh water snails as intermediate hosts for Schistosoma reproduction.
The worm eggs were first discovered in 1851 by the German pathologist Theodor Maximilian Bilharz during a post mortem procedure. Soon after the unusual morphology of Schistosoma was described. Schistosoma flukes are not hermaphroditic such as most other worms, but the sexes are separated into males and females. Female worms are smaller and live wrapped up by the larger male worm, who feeds her from his blood meals. The genus Schistosoma comprises of more than 20 species. Most relevant to causing Schistosomiasis in humans are S. mansoni, S. haematobium, S. japonicum, whose genomes have been sequenced a while ago.
Schistosomiasis is endemic in >70 mostly poor countries in Africa, Asia and South America, where it affects more than 200 million people. In fact, with about 200,000 annual deaths, it is the most deadly among all NTDs. The eggs hatch in water and the released miracidia need to penetrate the skin of a new host to survive, because they do not have a mouth for eating. This penetration can cause various forms of skin rashes, which are more or less intense depending on the immune status of the individual against Schistosoma antigens. The other symptom of an acute infection is called Katayama fever. It is caused by the travel of the trematode through the bloodstream into organs, mostly the liver. The worms develop through several stages into adult animals over a cause of several weeks.
If the infection is not treated, it will develop into a chronic infection stage. The main forms to be distinguished here are intestinal schistosomiasis and urogenital schistosomiasis. The type that will develop is species-dependent. S. mansoni and S. japonicum cause intestinal schistosomiasis where the large bowel and the rectum are enlarged by immune reactions against Schistosoma eggs, which can cause colon obstruction, blood loss and diarrhea. In contrast, S. haematobium causes urogenital schistosomiasis with symptoms such as heavy pain at urination and blood in urine, but also urinary tract infections. Female patients often also suffer from inflamation of the vagina and cervix, which can even lead to infertility.
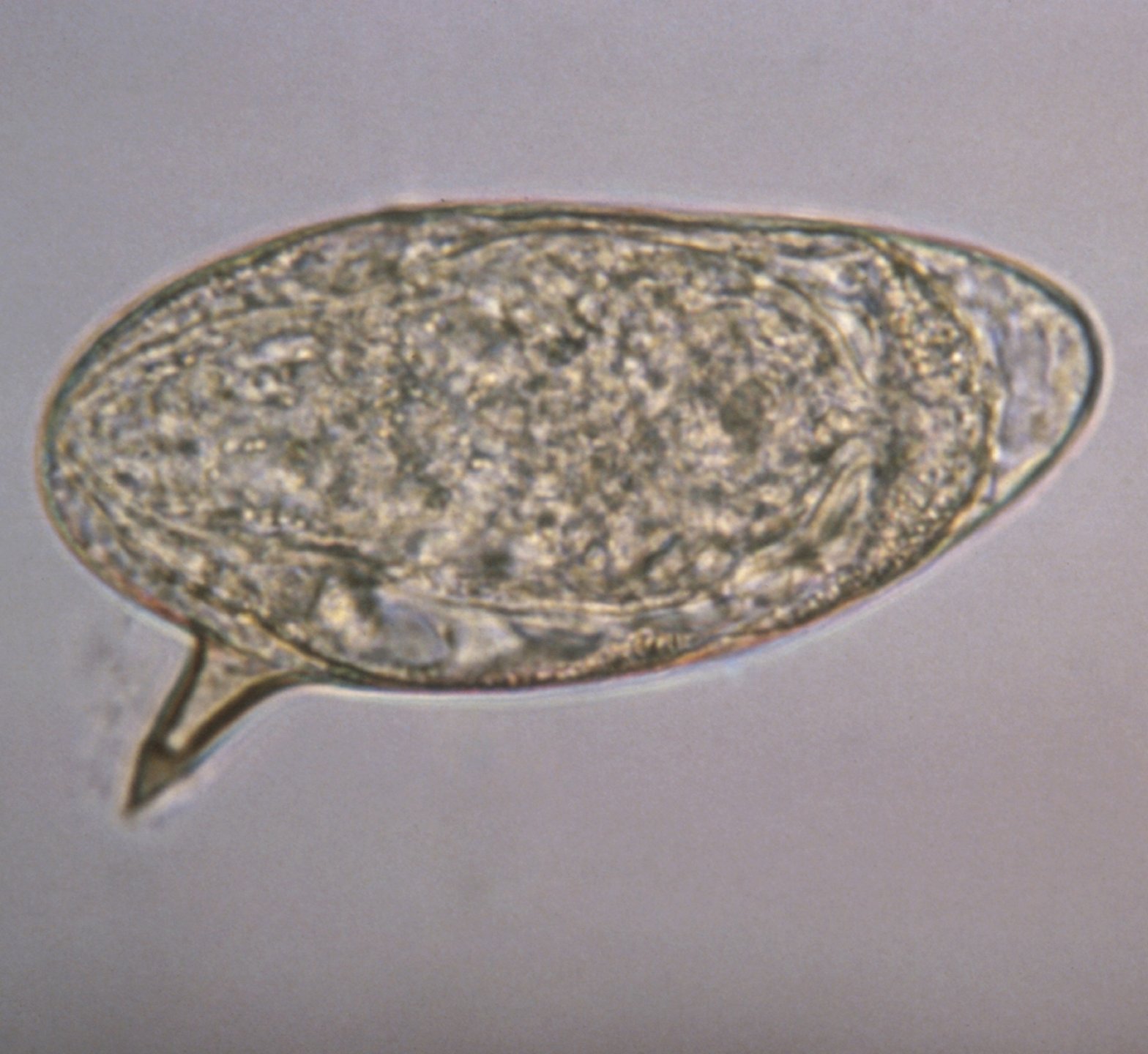
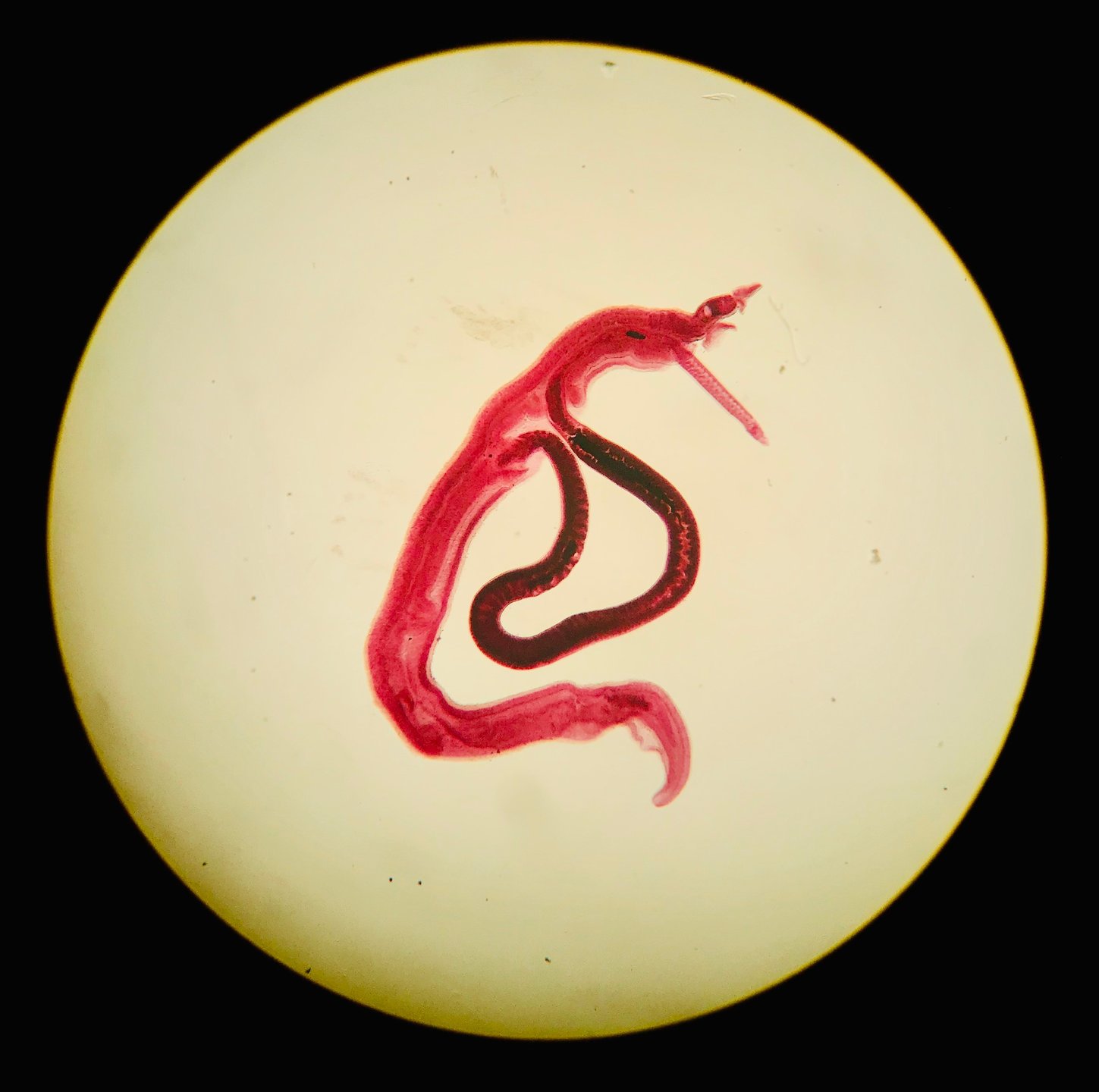
Molecular diagnosis
Schistosomiasis is traditionally diagnosed through microscopic techniques mostly enhanced through Kato-Katz enrichment. Immunological detection is possible through ELISA and lateral-flow tests. Yet, the probably most sensitive approaches to detect Schistosoma infections are molecular. The respective sample type is dependent on the type of schistosomiasis: fecal samples are used to detect intestinal schistosomiasis and urine or vaginal lavage is used to detect urogenital schistosomiasis. In case of negative but suspected positive patient, tissue biopsies may be examined in both types to find schistosoma eggs.
Molecular diagnosis of Schistosomiasis requires a sensitive extraction of DNA from urine and feces. Ideally, the DNA extraction protocol is field-friendly and can be applied in low-ressource setting laboratories or even literally in the field at the point-of-care. Amplification and detection of Schistosoma DNA itself is usually performed using real-time PCR, RPA, RAA or LAMP.
Applicable Xpedite Diagnostics products
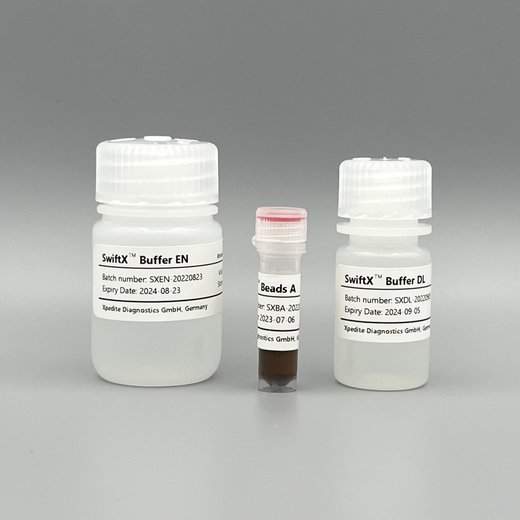
SwiftX™ DNA (25 extractions)
For magnetic bead-based capture of Schistosoma eggs and rapid extraction of Schistosoma DNA.
Validated for urine samples and cervicovaginal lavage.
Go to productUser protocols
Magnetic capturing of Schistosoma eggs from urine followed by DNA extraction
Rostron et al. (2019) Parasites & Vectors 12: 514 (see below for direct link)
- urine was defrosted and 100µL were taken for DNA extraction
- 200µL Buffer EN and 15µL Beads A were added and mixed
- mixture was incubated for 3 minutes for binding of Schistosoma eggs to Beads A
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A with Schistosoma eggs were resuspended in 100µL Buffer DL
- mixture was heated for 5 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time RPA detection
Magnetic capturing of Schistosoma eggs from urine followed by DNA extraction
Archer et al. (2020) Molecules 25: 4175 (see below for direct link)
- 50µl fresh or frozen urine was used for DNA extraction
- 100µL Buffer EN and 7.5µL Beads A were added and mixed
- mixture was incubated for 3 minutes for binding of Schistosoma eggs to Beads A
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A with Schistosoma eggs were resuspended in 100µL Buffer DL
- mixture was heated for 5 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time RPA detection
Centrifugation of urine for Schistosoma egg sedimentation followed by DNA extraction
Frimpong et al. (2021) Acta Topica 216: 105847 (see below for direct link)
- approximately 40mL urine was centrifuged for 10 minutes at 3000rpm
- supernatant was discarded
- pellet was resuspended in 100µL water
- 100µL Buffer DL and 15µL Beads A were added and mixed
- mixture was heated for 10 minutes at 95°C
- tubes were removed from heat block and placed in a magnetic rack for 1 minute
- supernatant was used for real-time RPA detection
Magnetic capturing of Schistosoma eggs from cervicovaginal lavage followed by DNA extraction
Archer et al. (2022) PLOS Neglected Tropical Diseases 16: e0010276 (see below for direct link)
- 50µl frozen cervicovaginal lavage was used for DNA extraction
- 100µL Buffer EN and 7.5µL Beads A were added and mixed by vortexing
- mixture was incubated for 3 minutes for binding of Schistosoma eggs to Beads A
- tubes were placed in a magnetic rack for 1 minute for bead separation
- supernatant was removed
- Beads A with Schistosoma eggs were resuspended in 100µL Buffer DL
- mixture was heated for 5 minutes at 95°C
- remove tubes from heat block and place in a magnetic rack for 1 minute
- supernatant was used for real-time RPA detection
Technical information
Documents and Videos
Scientific literature
References
Please note:
Most of the below publications reference the SpeedXtract® Nucleic Acid kit from Qiagen. This kit has been discontinued. The SwiftX™ DNA kit is identical in handling and performance to the former SpeedXtract kit. Thus, all published work can be reproduced using SwiftX™ DNA.